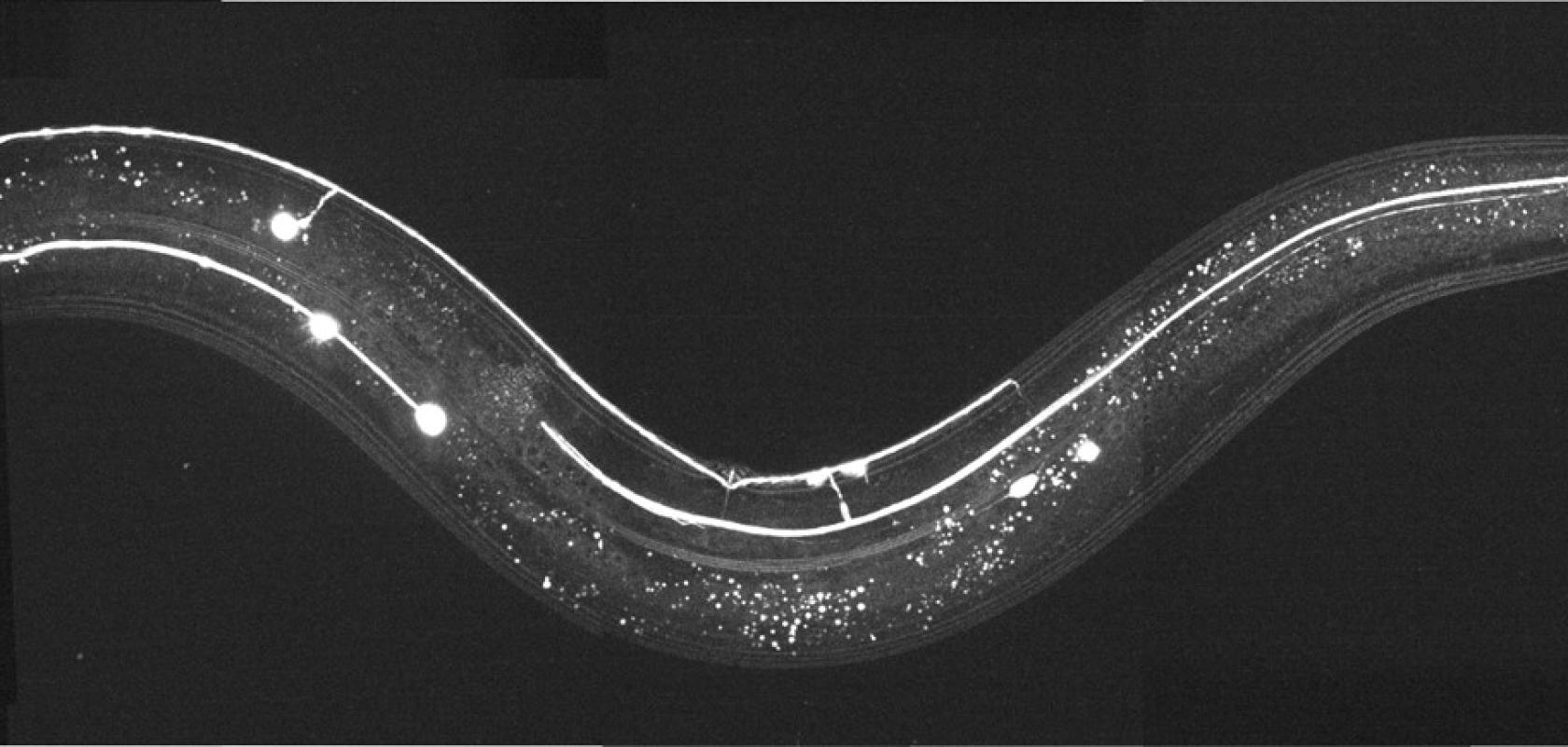
Bioluminescent microscopy’s unique advantages are pushing researchers to challenge it against conventional fluorescent methods. When detecting and imaging live cells and animals, the use of bioluminescence reporters has shown to be a promising method, especially in neuroscience research.
Dr. Michael Krieg, from the Institut de Ciencies Fotòniques, Castelldefels (ICFO), Spain, and his collaborators demonstrate volumetric imaging of fast cellular dynamics by exposing the advantages and pushing the limits of bioluminescence microscopy on Caenorhabditis elegans and other biological model organisms1.
The advantages and challenges of bioluminescence
Bioluminescence microscopy generates a signal by a chemical reaction of an enzyme and its substrate. It is similar to fluorescence microscopy but one significant advantage is that it does not use excitation light, which increases the specificity of the signal and avoids background signal from endogenous autofluorescence.
Another advantage is the reduced stress of the sample, which might induce some unwanted, potentially toxic effects in the sample.
Fluorescent markers also tend to bleach over time, limiting long-time observation of live samples. Those effects are efficiently mitigated using bioluminescence microscopy, which obtains the energy required for the signal generation from chemical processes rather than the photophysical process involved in fluorescence.
Nonetheless, a major drawback of bioluminescence is its rather low signal intensity, resulting in long exposure times and low SNR images in microscopy, limiting its applicability in dynamic processes, which are of big interest when observing living specimens.
The experimental setting
Dr. Krieg and his group optimised the experimental pipeline for bioluminescence. Using extremely photon-starved samples to challenge these limitations, they aimed to provide high SNR images at high temporal resolution. To achieve this, he used Nano Lanterns for the dyes, which are brighter and more varied in their spectral properties than the classically used luciferases. Since a standard widefield microscope did not provide any signal, they built a highly light-efficient microscope termed ‘LowLiteScope’.
Additionally, as the resulting images were still difficult to inspect due to the low signal available, content aware image restoration (CARE) was applied to improve these, which provided clear, high contrast images at millisecond exposure times.
Imaging with quantitative CMOS technology
To ensure efficient detection, Hamamatsu Photonics’ quantitative CMOS (qCMOS) camera ORCA-Quest was used to offer the best images from the available photons. Its extremely low readout noise of 0.27 e-, alongside its high quantum efficiency, provides the optimal signal-to-noise ratio down to almost the single photon level.
Apart from the raw sensitivity, another of its advantages lies within its geometrical design: in combination with the optics of the LowLiteScope, the pixel pitch of 4.6μm provides a perfect tradeoff between light collection efficiency and spatial resolution.
Since the ORCA-Quest delivers a pixel count of 9.4MP, there were no sacrifices with regards to the field of view. The pixel pitch and pixel count also benefited another evolution of the method: in order to increase the speed of 3D acquisitions, lightfield microscopy was also included. The small pixel sizes and the high pixel count of the ORCA-Quest allowed the projection of the 3D lightfield onto the two dimensional camera sensor, enabling rapid 3D imaging of the whole C. elegans using bioluminescence. Classical lightfield imaging requires a complicated computational reconstruction of the 3D object which might take up to 30 minutes. To speed up this process, a neural network was employed, reducing the time for 3D reconstruction down to 100ms. Using this scheme, satisfactory results can be achieved with exposure time down to five seconds. To further reduce the minimum exposure time in order to image sample dynamics in three dimensions, CARE was applied before the reconstruction. This reduced the achievable exposure times further up to a factor of 20, while maintaining acceptable exposure times.
Results and future applications
Luis Felipe Morales-Curiel and Dr. Michael Krieg have now established bioluminescence microscopy as a versatile addition to the biologist’s toolbox. Bioluminescence offers the benefits over fluorescence of lower stress to the samples as well as higher specificity. The common drawbacks of bioluminescence techniques, typically being low signal yield and resulting long exposure times were overcome by optimising the whole imaging pipeline. This included using the brightest bioluminescence dyes available, recording the data with instrumentation optimised for low light sensitivity, and finally processing the data with modern machine learning based approaches.
References
1 https://www.nature.com/articles/s42003-022-04292-x
2 Communications Biology | (2022)5:1330 | https://doi.org/10.1038/s42003-022-04292-x | www.nature.com/commsbio
Further information
For more information about Hamamatsu’s work in the life sciences sector, visit: www.hamamatsu.com