Just over four years ago, Dr Geraint Wilde, business manager for microscopy at Andor Technology, and colleagues, noticed a nuance in what their company's customers wanted. Many were using 'Dragonfly', a spinning disk confocal microscope system, to image single molecules, live cells and rapidly capture whole embryos and thick tissue in 3D at high-resolution – and even beyond the diffraction limit to super-resolution. But long-waiting times to access equipment in many core facilities around the world, were leaving scientists frustrated.
“These confocal systems were amongst the busiest in a facility, but from what we could see, quite a few researchers were using them for fairly routine 2-, 3-, 4-colour fluorescence imaging,” says Wilde. “Researchers still wanted confocal imaging, they wanted a high contrast image and they also wanted to get results quickly, so they were drawn to the Dragonfly as it is 10 times faster than your conventional confocal point scanning system.”
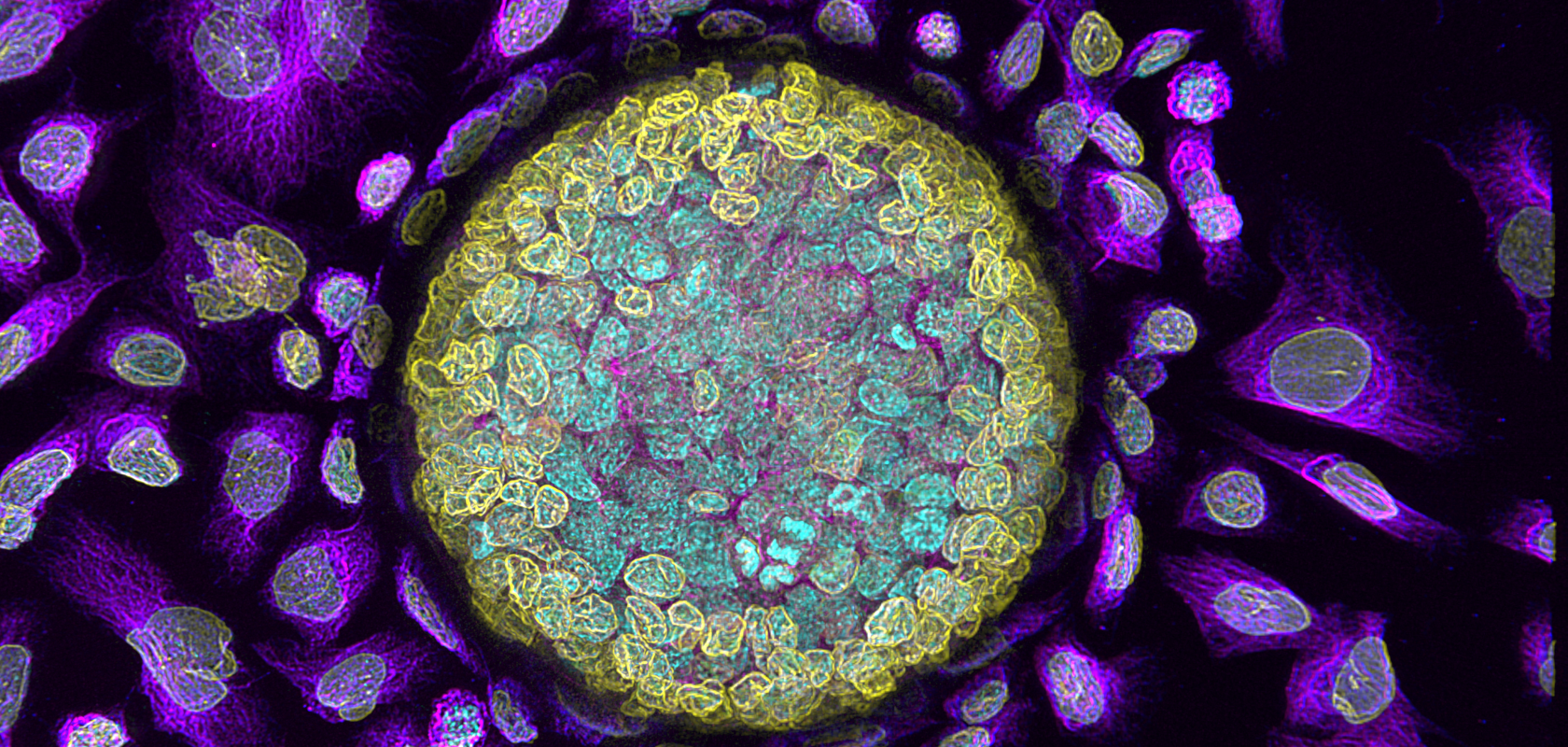
What's more, Wilde and colleagues had also noted that life science funders were awarding more and more grants for research that could demonstrate a clinical application in, say, diabetes, lung disease or cancer. Consequently, the demand for 3D imaging of model organisms such as Zebrafish, Drosophila fruit flies, the Caenorhabditis elegans nematode, or even an artificially created organoid, was rising. Given this, and the need to raise accessibility to confocal equipment, the Andor team developed an easy-to-use benchtop high-speed Nipkow disk microscope system – BC 43 – that could provide high-quality 3D imaging to more researchers.
“The system really is simple and you can see the detail that you would in a sophisticated confocal system,” highlights Wilde.
Shrinking the system
The BC43 launched late last year with the chief challenge being to shrink the Dragonfly platform from some 1,500cm by 70 x 70cm to a more compact 50cm by 45 x 50cm. Key steps were to re-think the system's illumination set-up and maintain the functionality of a traditional microscope.
According to Wilde, the Dragonfly laser module comprised up to eight lasers, including diode-pumped solid state devices, and was housed in up to two modules, each the size of a tower PC workstation. However, he and colleagues decided to use four lasers, instead of the previous eight, which was deemed sufficient for routine imaging. They also switched from DPSS lasers to laser diodes, and as Wilde says: “In doing this, we brought the laser module down to the size of a thick, hardback book.”
Wilde and colleagues also opted for a novel but relatively simple brightfield illumination set-up in which contrast was created through a software algorithm, rather than conventional phase contrast or differential interference contrast (DIC) with the myriad associated optics. The benchtop system also uses a single, small scientific CMOS camera, instead of the two cameras in its original platform, and has a specially-developed compact sample stage.
“We investigated illuminating the sample with LEDs at different angles and then used software to rationalise this set-up and provide a DIC-type image,” points out Wilde. “With software we could rationalise the brightfield, take away the eyepieces by adding automated hardware control features, strip away the focusing knobs – there's only two buttons to push – and really reduce the component-count to simplify the user experience.”

The BC43, a desktop confocal microscope from Andor
Along the way, the Andor team simplified the Dragonfly's control software for the BC43, whilst still enabling their “vision for non-experts to capture multi-colour 3D images,” as Wilde says. The company's 3D visualisation rendering engine within 'Imaris' was also embedded in the acquisition software, and users can now see their 3D sample develop in front of their very eyes as the BC43 works its way through a sample volume.
Now the platform is delivered, Wilde is adamant that users do not face any real compromise during routine imaging. Resolution is close to that of the Dragonfly while image acquisition remains fast, coming in some six times faster than traditional confocal imaging. “Obviously with only four lasers you have less flexibility on the fluorophores you can use, but the most popular ones are covered,” says Wilde. “Also, the BC43 has no additional ports, so you can't bolt on other tools for, say, optogenetics. This remains the domain of the Dragonfly.”
So far, the system has been well-received, with systems installed at the National Cancer Institute in Amsterdam, small biotech companies, as well as medical school laboratories in several countries including Japan and Denmark. And as Wilde and colleagues had envisaged, users have been tracking embryo development and division, as well as imaging organoids and large tissue sections, including parts of the brain.
“We get this 'wow', especially from researchers that have been using larger, more conventional confocal microscope systems,” says Wilde. “When we install a system, we'll load in a sample and our customer will say let's go for a cup of tea [while data is being acquired]. But by the time we've had this conversation, we've got the image.”
Unsurprisingly, the entire microscopy community is seeing a rise in the development of more accessible microscope systems, with the likes of Leica, Thermo Fisher Scientific, Bruker and Renishaw providing a vast range of imaging modes including optical, Raman, FTIR and scanning electron microscopy. In 2017, Delong Instruments delivered 'the world's first benchtop transmission electron microscope (TEM)' and now provides a series of compact all-in-one instruments that provide TEM, scanning transmission microscopy, electron diffraction and, most recently, energy dispersive spectroscopy.
“When we started out with BC43, we didn't have a like-for-like competitor,” says Wilde. “But many other companies have also seen there is this demand and we are seeing more benchtop systems.”
“Our aim was to make confocal microscopy more accessible,” he adds. “And what we are seeing now is that facilities that felt confocal technology was out of their reach financially can now access this technology.”
Start-ups and benchtop systems
Still, it's not just the established industry players that are stepping up to accessibility – a raft of start-ups is developing more affordable analytical systems. Earlier this year, US-based super-resolution microscopy start-up Eikon, co-founded by Nobel Laureate, Eric Betzig, won a mighty $518 million to develop commercial systems to track proteins moving in cells in real time with, as the company says, 'exceptional spatial and temporal resolution'. At around the same time, ONI, a UK-based biotech start-up, won $75 million to further commercialise its super-resolution systems. The company's desktop 'Nanoimager' is described as a complete package for super resolution microscopy, and is already being used in myriad research organisations including Weill Cornell Medicine, Cancer Research UK and AstraZeneca.
One start-up that is taking a different approach is the newly-formed, Ireland-based turboTEM, headed up by Professor Lewys Jones, also leader of the Ultramicroscopy Group at Trinity College Dublin (TCD). In the last few years, Jones, and TCD colleagues, have been developing hardware and software to make scanning transmission electron microscopes (STEMs) more cost effective, easier-to-use, longer-lasting, and ultimately, more accessible to researchers around the world.
In a first venture, the TCD team developed a digital electron-counting system, called Pulse, for annular dark-field STEM imaging. Efficient electron detection is critical to reduce the electron dose needed to image a sample, and improve the speed and precision of STEM signals, but buying a new conventional electron counting sensor isn’t cheap. However, with Pulse, Jones and colleagues have developed a custom PCB package that can plug directly into a microscope's existing detector to enable single electron counting.
The system has already been trialled on several scan controllers including the Gatan Digiscan II as well as with market-leading STEM detectors from JEOL, Nion, and Thermo Fisher. “We've made a big effort to use, say, standard connectors and TTL [Transistor-Transistor Logic] signal triggering so it can work with any machine,” highlights Jones.
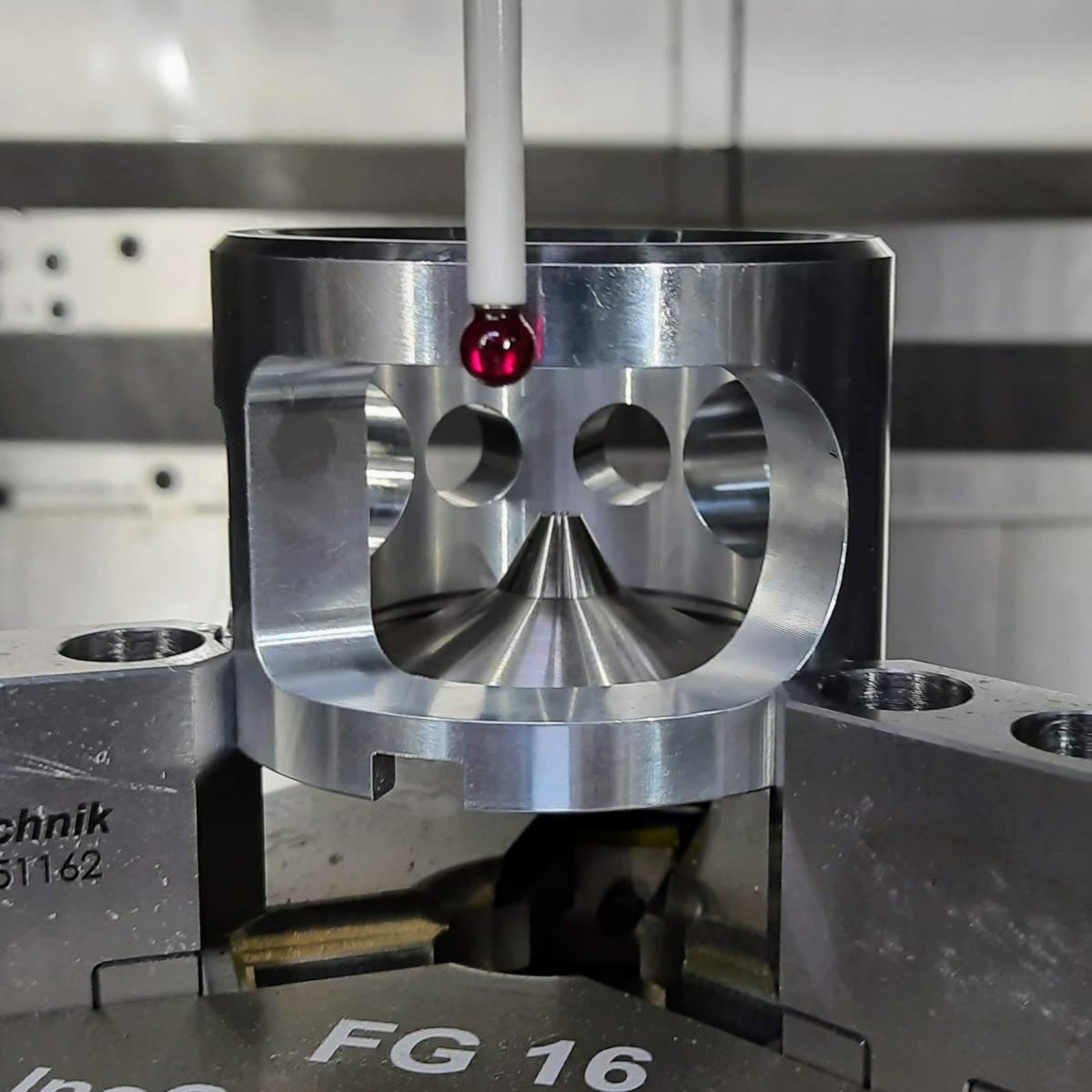
A component of the user adjustable pole-piece from turboTEM as it undergoes measurement to ensure dimensional accuracy using a high-precision coordinate measuring machine. Image: turboTEM
The turboTEM director expects the typical Pulse user to perhaps have a five to ten year old machine that needs upgrading for higher speed or lower dose imaging. Alternatively, he or she could be a researcher with a brand new instrument who simply wants the absolute best possible performance. Either way, he and turboTEM colleagues will be designing, assembling and testing all their devices in Ireland, with the first unit scheduled to be shipped in the next few months.
“We are an embryonic company but are also working with Point Electronic [an independent supplier of detectors, acquisition and control systems for electron microscopes] to distribute our devices globally,” says Jones. “We've had enquiries from National Labs across several continents.”
Excitingly, as the fledgling company ramps up manufacturing of its digital electron counting system, it is also poised to launch a second device – an adjustable gap pole piece for the objective lens of a TEM. In any shared facility, the electron microscope's objective lens pole-piece will be set to a certain application, which may not suit everyone on the long waiting list of users. For example, small pole-gaps can provide the lowest lens aberrations for high resolution microscopy. Meanwhile larger gap pole-pieces provide more room for in-situ experiments with larger sample set-ups, but won't reach the highest optical performance.
Given a flagship TEM can cost in the region of $5 million, having a less than satisfactory set-up for many users is hardly ideal. So with this in mind, Jones and TCD colleagues have spent some four years designing, 3D printing, prototyping and building a replacement pole-piece for electron microscope platforms with the JEOL 200/300 kV architecture. With a sliding concentric mechanism, the pole-piece can be adjusted by the user without engineer support while maintaining the all-important vacuum in the electron microscope column.

A screw thread is tapped in the body of turboTEM’s user adjustable pole-piece, ready for a protective cover plate to be bolted on
Initial community feedback has been incredibly enthusiastic with researchers far and wide highlighting how the pole-piece will make microscopes more flexible and versatile. As one potential user put it: “The new concept of the user-adjustable pole-piece will free microscopists from the chronic pain of pole-piece selection.”
Jones himself is excited – the turboTEM team has just assembled their first unit for a research partner, and this is currently undergoing quality control and accelerated-wear lifetime analysis. “We expect it to easily outlast the instrument that it has been designed for,” he says.
And of course, for Jones and colleagues, this sentiment of durability with flexibility, is a huge part of what their research has been about. Pointing to today's microscopes, which may run into operational problems if today's energy crisis leads to rolling brownouts, he highlights: “Re-starting these instruments can take a week – they really need to be more resilient and a lot less frail than they are today.”
When it comes to re-designing an existing technology, Jones and colleagues have always ensured they keep a close eye on systems being widely used in other, related industries. A case in point is medical detectors. Right now, electron microscope detectors are expensive and only available from a handful of manufacturers. But if the turboTEM colleagues could re-purpose a microscope detector from an analogous system in the medical industry – which is orders of magnitude larger than the microscopy community – then they could draw on the economies of scale that already exist here and deliver a more affordable option.
“We currently have two students in our research group that are looking at low-cost detector technologies,” he says. “It's not always obvious [what to do], as we're working with electrons and not light, but if there's ways to modify other systems for the world of electron microscopy, this can reduce costs.”
“We are always looking to repurpose [devices] from adjoining technologies... and I wouldn't be surprised if, in a couple of years from now, we'll have also developed low-cost electron microscope detectors,” he adds.
Sponsored: The ever-expanding palette of colours for multiplexed fluorescence detection
The capacity to simultaneously identify and localise multiple target biomolecules in complex, heterogenous specimens has long been one of the primary attributes driving applications of fluorescence microscopy in the biological sciences.
The ultimate upper limit of “multiple” is dictated by biological imperatives. It is at least 10,000, which is approximately the number of non-identical proteins in a typical mammalian cell, all of which interact in some way to generate structure and function.
Multiplex detection schemes for fluorescence microscopy are usually based on spectral encoding, where the excitation and emission spectral characteristics of fluorescent dye-labelled antibodies and oligonucleotides or fluorescent proteins (FP) encode the identity of target biomolecules to which they are attached.
The scope of these methods is commonly limited to about five targets due to the spectral overlap between fluorescent dyes and fluorescent proteins (FP), which can result in target misidentification due to signal crosstalk. Increases in the number of resolvable targets have been achieved through improvements of illumination sources, optical filters, and spectral unmixing algorithms1.
Careful optimisation of optical filters and secondary antibody labelling reagents, together with application of a robust unmixing algorithm which corrects raw fluorescence image data for crosstalk as well as nonspecific labelling, 10-fold multiplex imaging of brain tissue has been demonstrated2.
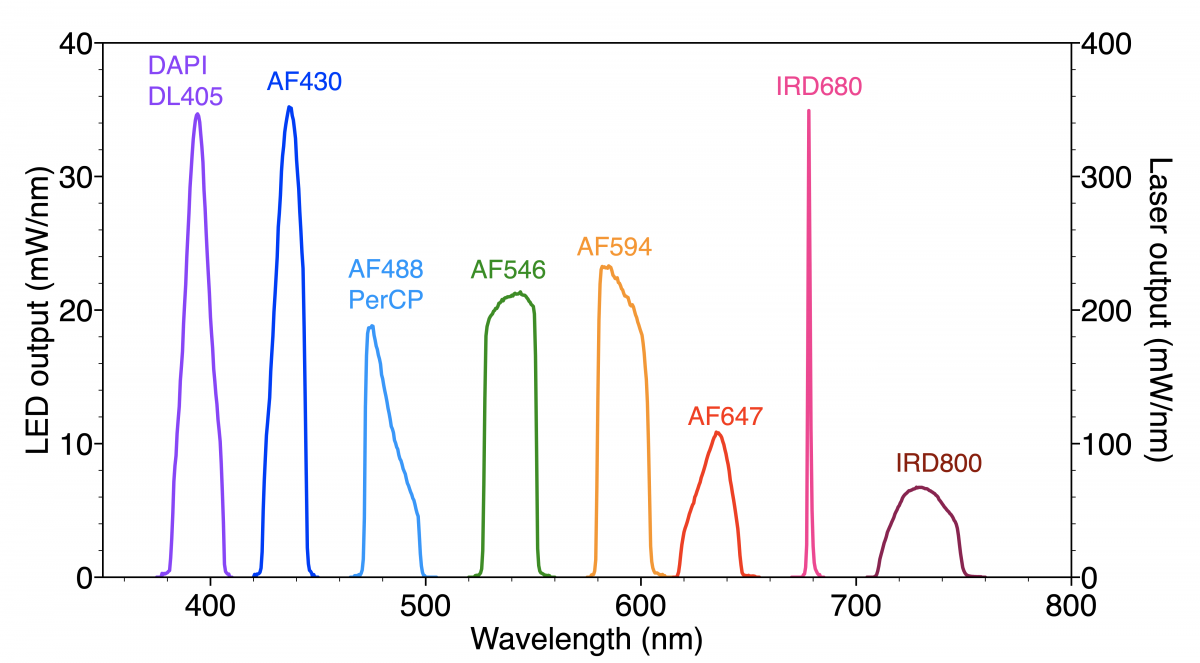
Figure 1: Spectral output of Spectra Light Engine optimised for selective excitation of fluorescent dyes used for 10-plex immunofluorescence detection
The spectral output of the solid-state Light Engine optimised for selective excitation of the panel of fluorescent dyes used in this protocol is shown in Figure 1. The Spectra Light Engine (Lumencor) incorporates a proprietary mix of solid-state light sources based on three distinct technologies, namely LEDs, light pipes and diode lasers, to achieve the required distribution of spectral outputs.
Precise bandpass filtering within the Light Engine contributes to the spectral purity and selectivity required for such advanced multiplexed fluorescence analyses. The level of multiplexing can be extended by successive rounds of stripping, re-labeling and imaging, using the same fluorescent dye panel and filter configuration. With five cycles of stripping, 10-plex re-labeling and imaging, simultaneous localisation of 50 biomarker targets in whole rat brain coronal sections has been demonstrated2.
Multiplexing technologies capable of simultaneously identifying and localising hundreds and thousands of biomolecular targets have been developed through the application of combinatorial labelling techniques. For example, six spectrally distinct fluorescent dyes can be assembled in 15 unique binary combinations3. MERFISH (multiplexed error-robust fluorescence in situ hybridisation) is a massively multiplexed single-molecule imaging technology capable of simultaneously measuring the copy number and spatial distribution of hundreds to thousands of target RNA species in single cells4,5,6.
In MERFISH, target-associated barcodes are read iteratively. First, encoding oligonucleotide probes are hybridised to target RNAs. Then fluorescent dye-labelled readout oligonucleotide probes are successively added, spatially localised at the single-molecule level, and then extinguished. Readout probes must hybridise exclusively to encoding probes, and not to cellular nucleic acids. The encoding probe identity is accumulated across successive imaging cycles, with binary bit value '1' assigned if the encoding probe is detected by hybridisation of a readout probe, and a '0' value if it is not. In principle, 12 hybridisation+imaging cycles would be required to identify 4095 (212-1) unique encoding probes.
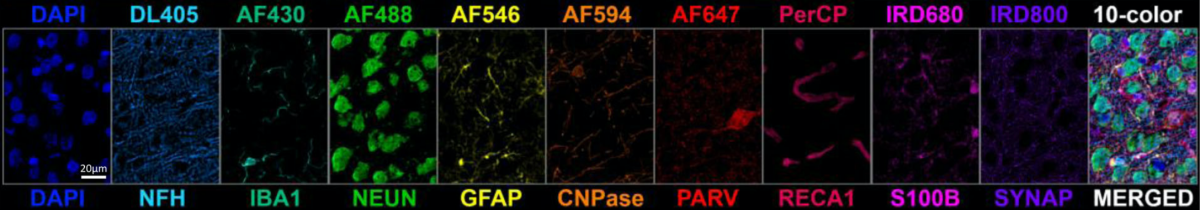
Multiplexed detection of nine neural cell biomarkers plus DAPI (nuclear stain) in rat brain sections. Reproduced from [2] under CC 4.0
In practice, the binary coding space is underpopulated to minimise false negative (1>0) and false positive (0>1) calling errors and consequent misidentification of target RNAs. For example, identification of 10,050 RNA targets using a 69-bit barcode has been demonstrated6. The number of hybridisation+imaging cycles, and therefore the time required to complete data acquisition, can be reduced by simultaneous detection of readout probes with spectrally distinct fluorescent labels in each cycle (e.g. 23 imaging cycles x 3 fluorescent labels = 69 bits)6. Strategies to fulfil the promise of sensitive, accurate highly-multiplexed bioanalyses for large target number samples typically focus primarily on the biochemistry of labelling.
Moreover, such complex bioanalyses necessitate precise illumination. Solid-state Light Engines like the one described here from Lumencor. ensure fidelity as well as consistency for fluorescence detection in complicated labelling techniques. Spectral specificity, spectral breadth and high optical power are each required of solid-state Light Engines which must also deliver stability and longevity to support best data quality, in terms of both fidelity and extended multiplicity.
References
1 J Seo, Y Sim, JB Chang et al. PICASSO allows ultra-multiplexed fluorescence imaging of spatially overlapping proteins without reference spectra measurements. Nat Commun (2022) 13:2475.
2 D Maric, J Jahanipour, B Roysam et al. Whole-brain tissue mapping toolkit using large-scale highly multiplexed immunofluorescence imaging and deep neural networks. Nat Commun (2021)12:1550.
3 AM Valm, JL Mark Welch, GG Borisy. CLASI-FISH: principles of combinatorial labelling and spectral imaging. Syst Appl Microbiol (2012) 35:496–502.
4 KH Chen, AN Boettiger, X Zhuang et al., Spatially resolved, highly multiplexed RNA profiling in single cells Science (2015) 348:aaa6090.
5 JR Moffitt, J Hao, X Zhuang et al., High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc Natl Acad Sci U S A (2016) 113:11046–11051.
6 C Xia, J Fan, G Emanuel, J Hao, X Zhuang et al., Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression Proc Natl Acad Sci U S A (2019) 116:19490–19499.