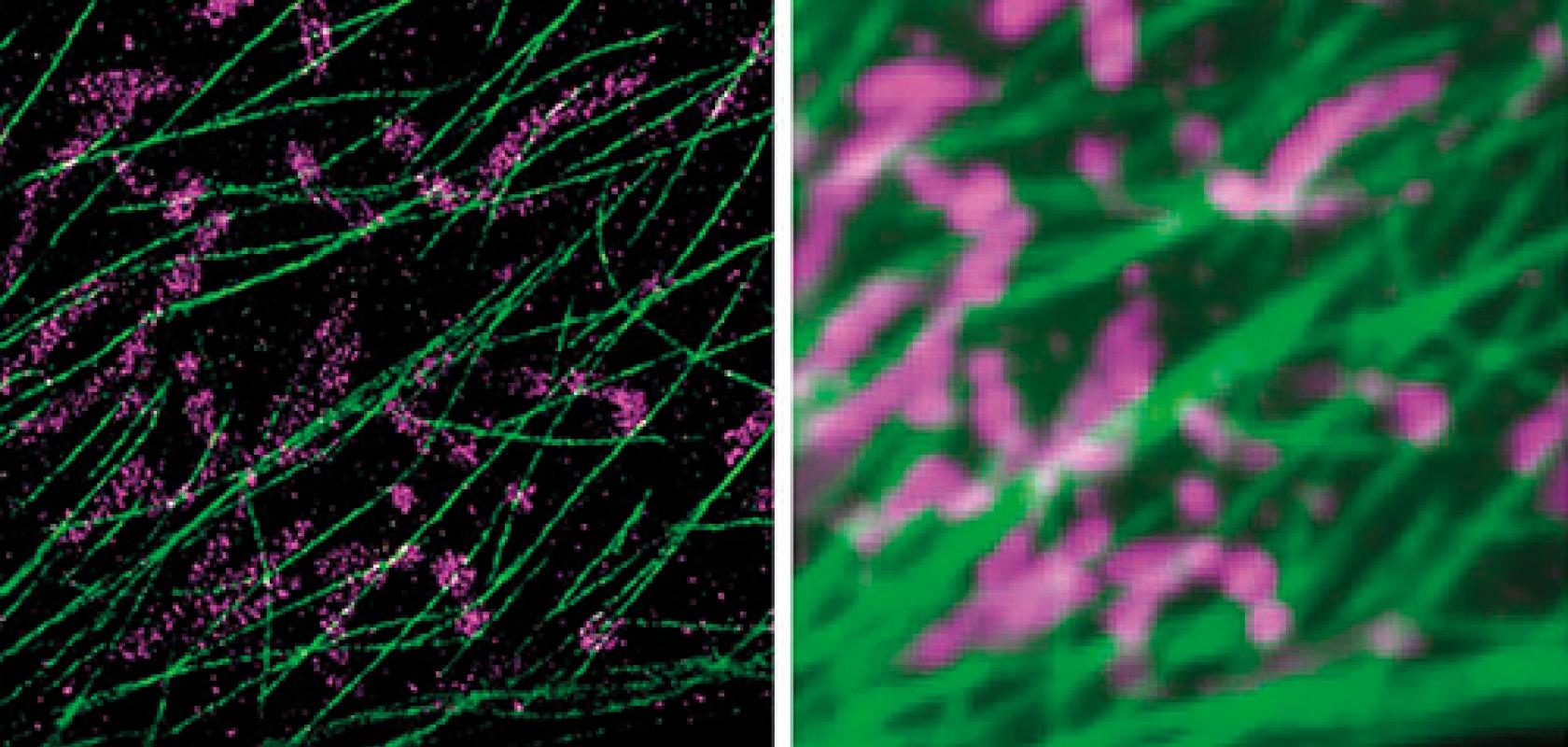
DNA-PAINT and Exchange-PAINT technologies (right) improve the resolution abilities of single-molecule microscopes (left)
The commercial introduction of super-resolution microscopes, which allow scientists to observe cellular processes at a scale smaller than the 200nm diffraction limit of light, has been one of the most significant developments in the field of microscopy in decades. However, the price point of this advanced technology often means that it is restricted to large research facilities and big companies.
Thanks to developments made by both research groups and commercial organisations, super-resolution techniques are becoming cheaper and more accessible to biologists.
For example, scientists at Harvard University’s Wyss Institute have developed a relatively inexpensive microscopy platform that can achieve an optical resolution of less than 5nm, the size of a small protein. The new set of methods can reveal features in single molecules, using conventional microscopes found in most laboratories.
In addition, super-resolution pioneer Leica has released a solution that allows a confocal microscope to capture super-resolution images, and start-ups LIG Nanowise and Oxford Nanoimaging have launched desktop systems that lower the cost of super-resolution imaging.
Nanometre optical resolution
Despite super-resolution microscopes being able to visualise at 10-20nm resolution, this is still not powerful enough to distinguish between individual features within densely packed protein complexes, according to researchers from Harvard’s Wyss Institute for Biologically Inspired Engineering.
With a new set of methods developed at the Wyss Institute, however, scientists will be able to observe and track individual proteins within molecular complexes without needing to spatially resolve them, and without requiring an expensive large super-resolution microscope.
‘Proteins do not usually work in isolation but in larger complexes that enable cells to communicate with each other, move cargo around in their interiors, and replicate their DNA,’ said Wyss Institute core faculty member Professor Peng Yin. ‘Our ability to observe and track each individual protein within these machines is crucial to our ultimate understanding of these processes.’
Reporting in Nature Nanotechnology1 in 2016, Professor Yin and his team describe the discrete molecular imaging (DMI) technique, which builds upon their super-resolution microscopy platform, DNA points accumulation for imaging in nanoscale tomography (DNA-PAINT).
‘DNA-PAINT uses transient binding between two complementary DNA molecules – one that is attached to the molecular target of interest, one that is floating in the solution and conjugated to a fluorescent dye. When they bind together, they create transient blinking events at the molecular target site that are visible through the microscope to create super-resolution microscopy images,’ explained Mingjie Dai, first author of the work.
The more recent and complementary qPAINT method – which stands for quantitative PAINT – allows researchers to quantitatively count the number of molecular features within large molecular complexes. ‘Now, with DMI, we are adding the extra power of being able to resolve every single molecular feature that are on the protein size, which is about 5nm in size,’ Dai added. ‘Protein-protein interactions are the foundations of biological life – visualising them in the complex is only the first step in trying to potentially manipulate them on a single molecule level,’ Dai said.
‘Ultimately, new DMI technology provides researchers with a way to study – and later possibly diagnose – certain diseases based on individual molecular signatures,’ added Marcin Barszczewski of Andor, which provided its Andor Zyla 4.2 and iXon Ultra 897 cameras for the group’s research.
DMI also complements current structural biology methods like x-ray crystallography and cryo-electron microscopy, and provides an easy, fast and multiplexed method for the structural analysis of many samples in parallel.
Ultivue, a start-up launched out of the Wyss Institute, is focused on developing imaging solutions to unfold new areas of biology by using DNA-PAINT to co-localise an unlimited number of targets.
Lens on cost
Earlier this year, LIG Nanowise, a Manchester, UK-based nanotechnology start-up, launched a device based on super-resolution microsphere amplified lens (SMAL) technology, which lowers the cost of super-resolution imaging. With a magnification of x400, the lens allows scientists to image at an optical resolution of 70nm using standard optical microscopes.
According to LIG Nanowise, the system costs five times less than stimulated emission depletion (STED) microscopy and stochastic optical reconstruction microscopy (STORM) systems, and is estimated to be 10 times cheaper than a standard electron microscope.
The SMAL technology was developed by Professor Lin Li, director of the laser processing research centre at the University of Manchester, who demonstrated the basic principal of this technique in 2011.

The Nanopsis nanoscope can image with a 70nm resolution
LIG Nanowise has deployed this technology inside its new Nanopsis imaging system, which the company describes as the ‘world’s first’ microsphere nanoscope. It works by using a microsphere – tiny transparent spherical beads – to gather sub-wavelength light and convert it into a virtual super-resolution image. Software then stitches these images together in real time to generate full colour, widefield scans of materials and life samples – resolving details down to 70nm.
‘Researchers can use our microscopes to validate samples and carry out routine work in their own laboratory without having to waste valuable time booking into an imaging centre.
This is because unlike other super-resolution technologies, which require a huge amount of expertise, our Nanopsis nanoscopes can be used by anyone with basic undergraduate scientific training – making it fast, convenient and highly cost effective,’ explained Professor Li, who is also chairman of LIG Nanowise.
‘Equally important is the fact that these reliable, repeatable imaging results are delivered at the frontline of research, rather than part of a disjointed process in an inaccessible centre. Our aim is to make super-resolution imaging more accessible to researchers across the globe,’ he added.
Another company marketing a desktop instrument is Oxford Nanoimaging (ONI), which in 2016 started selling a super-resolution microscope capable of imaging the chemical reactions of individual molecules in real time.
The Nanoimager has the footprint of a desktop computer, is about 30 times smaller and significantly less expensive than current super-resolution microscopes. During development of the instrument, ONI raised £1.2 million in a seed funding round from investor Oxford Sciences Innovation.
The microscope – developed by a team led by Professor Achillefs Kapanidis and PhD student, Bo Jing at Oxford’s Department of Physics – uses rapid imaging of single fluorescent molecules and software to generate images that differentiate objects spaced as close as 20nm.

Imperial College researchers use the ONI Nanoimager (foreground black box), which has a 20nm optical resolution
‘The new microscope will take single-molecule imaging out of physics labs and centralised facilities, and into the hands of the chemist, the biologist, the biotechnologist,’ Professor Kapanidis said.
‘Scientists who don’t have a lab equipped with specialist physics kit and a large budget can still access the information this very powerful technology provides,’ added Oxford Nanoimaging CEO Jeremy Warren.
Confocal super resolution
In addition to advanced systems that offer resolutions down to 30nm, Leica Microsystems, one of the first companies to release a commercial super resolution system, has launched a solution that transforms a confocal microscope into a super-resolution microscope.
The HyVolution system, which was developed with image processing firm Scientific Volume Imaging (SVI), allows biologists to image multiple fluorophores simultaneously and capture intracellular details with resolution down to 140nm.
Unlike super-resolution systems using a single external detection channel, HyVolution integrates with all confocal imaging modalities. Therefore, users can record all five channels simultaneously – without the need for sequential scanning, which slows the acquisition speed.
The solution merges optical super-resolution and computational super-resolution; the optical part is provided by confocal microscopy, and the computational part by deconvolution software.
SVI’s software, Huygens, can be applied to TCS SP8 confocal images directly after, or during, the acquisition stage. The deconvolution provides a significant contrast and resolution increase in acquired microscopy images. In combination with a small confocal pinhole, the Huygens deconvolution can provide a resolution improvement well below the conventional confocal diffraction limit.
Deconvolution also allows users to image with lower excitation power and faster scanning speed, thereby reducing sample damaging, photo-bleaching and photo-toxicity.
Reference: 1. Mingjie Dai, Ralf Jungmann, Peng Yin. ‘Optical imaging of individual biomolecules in densely packed clusters’ Nature Nanotechnology, 2016; DOI: 10.1038/nnano.2016.95
In January 2017, a Horizon 2020-funded project was launched to train experts on the use of light sheet microscopy (LSM) in order to advance cardiovascular research. The €1.5 million, four-year project will provide a comprehensive range of training to early stage researchers in the fields of biology, microscopy, optics and bio-computing to ascertain how a heart is formed and is able to regenerate upon injury. Six pre-doctoral researchers will be supported by the 4DHeart programme (4D analysis of heart development and regeneration using advanced light microscopy) in universities across Europe.
Cardiovascular (CV) disease is the leading cause of death worldwide. During adulthood, ischemic heart disease leads to heart failure and perinatal, congenital heart defects are found in more than 20 per cent of deaths, according to statistics referred to by 4DHeart. Moreover, genetic or epigenetic factors altering development can have an impact much later in life.
These facts underscore the need for a better understanding of the genetic and environmental factors that influence CV development. An important way to increase knowledge is by visualising cardiac development in vivo. Recent advances in microscopy mean monitoring CV development is possible at a cellular level in organisms such as the zebrafish model. Particularly revolutionary has been the development of light sheet microscopy (LSM).
The project will aim to exploit LSM for in vivo manipulation of cells in the embryonic zebrafish heart and measure with high precision biophysical parameters, by introducing novel features to LSM such as optical tweezers. High throughput cardiac imaging protocols for zebrafish larvae suitable for screenings will be set up. The aim is to generate a toolbox to be implemented into existing software packages allowing the zebrafish cardiac morphogenesis to be modelled.
The business partners in 4DHeart include Acquifer, Leica Microsystems, Philips Ibérica (a product division of Royal Philips Electronics), and Bitplane, a specialist in interactive microscopy imaging. The project is being coordinated by Professor Miguel Torres from the Centro Nacional de Investigaciones Cardiovasculares Carlos III.