Even as you read this, you are ageing with every single word – so medical technology’s promise of extending our healthy lives is greatly welcome. And it’s more than a promise – in most countries the average age is increasing. But that brings more ailments with it.
Cancer is right at the top of the list and, today, diagnosis is often a frustratingly slow process, observed Mikko Malmivaara, marketing director at Finnish microscope producer Grundium. ‘They cut open a patient and take a biopsy sample of tissue,’ Malmivaara explained. ‘Then the patient gets sewn up again and sent home.’ Meanwhile, technicians prepare a tissue sample and put it on slides and send them to a pathologist to study under a microscope for diagnosis. ‘They could be in a different city or even in a different country. Also, it is very common that the pathologist wants a second opinion, sending slides on to a colleague who might be in a different location again. This process might take up weeks.’
Therefore, Grundium has developed the Ocus line of small digital microscopes that enable telepathology. Instead of shipping the slides, or making a pathologist travel to a hospital where a patient is, Ocus scans slides and produces a digital image. Experts can then access it wherever they are, as long as they’ve got an internet connection. ‘They can do the diagnosis immediately while the patient is still in surgery,’ Malmivaara said. ‘You can share the entire user interface of our scanner between a number of pathologists. They can look at the sample at the same time, they can all use the controls, zoom in and out and move between different areas of the scanned image.’
Ocus is just one of many new miniature microscopes that are improving healthcare by enabling diagnosis nearer to patients. And that can span many diseases, not just those that emerge as people in wealthy countries age. Key to this trend is the ability to combine existing optical components with powerful algorithms. Meanwhile, the computing power and AI available also enables some developers to adopt innovative optical approaches.

The Grundium Ocus reconstructs images pixel by pixel to produce sharp images for remote viewing
Malmivaara stressed that digital pathology itself has been around for decades, based on multislide scanners, which range in size from the equivalent of a microwave oven to a large refrigerator. ‘Buying a high-level microscope, and then buying a multi-slide scanner, we’re talking up to a million dollars,’ he said. Whereas, they can scan up to 1,000 slides per time, Grundium’s ex-Nokia engineers have designed the Ocus to scan just one slide, making it small, practical and portable. ‘We’re not really competing directly with these big brands, but we are complementing their offering,’ Malmivaara said. ‘It’s so simple to use that anybody can be trained to use it in 15 minutes.’ The Ocus ‘costs about as much as a good microscope’ on its own, he added, and focuses on tissue and fluid applications for human, animal and plant life alike.
Grundium delivers the resulting images with the same kind of objectives and other optics as conventional microscopes – but the algorithms it uses to process those images are vital, Malmivaara explained.
‘We’re scanning pixel by pixel, and then stitching that up to an image that can be zoomed in and out of freely, and it looks perfect,’ he added.
These capabilities are already exploited worldwide by laboratories and hospitals including Memorial Sloan Kettering cancer hospital in New York, and Karolinska Institutet medical university in Stockholm.
Better diagnosis for all
Surprisingly, these elite laboratories are unified with medics working in rural Africa in optimising small, dedicated microscopes. That’s thanks to the Inclusive diagnostics for poverty-related parasitic diseases (Inspired) team at Delft University of Technology, in The Netherlands. The team has developed a microscope to diagnose the parasitic disease schistosomiasis.
One of them, Tope Agbana, said: ‘What we aim to achieve with the schistoscope is a point-of-care device with no need for technical expertise.
‘We can get into rural areas where real experts for manual microscopes are not available. We try to reduce the dependence on electricity, by integrating power. We try to integrate algorithms with the optics to automate the diagnostic process.’
The schistoscope builds on previous research the team had done to adapt a smartphone camera to become a microscope to detect malaria parasites. The team optimised the optical train to realise 8x magnification with a ball lens, sufficient to detect the parasites in blood samples. This work was not wholly successful, Agbana admitted. ‘The malaria parasite is one micron, really very small,’ he said. ‘You need to look at close to a million red blood cells before you can do a conclusive diagnosis. You cannot do that from the smartphone-based method. In our research lab here in Delft, it works. But when you get to the field, with all the dust and all the complication, it will not be the best option.’
Those lessons proved useful in schistosomiasis, where the parasite is 120 times bigger than in malaria. Agbana and colleagues moved away from the design constraints imposed by smartphones to a Raspberry Pi.

The schistoscope was originally designed to detect parasites with smartphones, but has now evolved to a system exploiting the Raspberry Pi
‘You can build your optics from scratch, and then use the Raspberry Pi’s computational capacity to drive your algorithm to detect the object in your acquired image,’ he explained. The resulting device uses an algorithm to recognise the coconut shape of the schistosome egg.
The Delft team is working with its Netherlands compatriots at Leiden University, as well as universities and research centres in Nigeria and Gabon. The researchers had been due to be testing the schistoscope in Gabon before the pandemic.
‘We continue developing our product in parallel,’ Agbana said. ‘We shipped schistoscope 3.0 to Gabon three months ago. Based on the feedback we got from the medical team on the ground, we have now optimised on that iteration.’
The collaboration is exploring possibilities to raise funds for its rollout. ‘Hopefully, by next year we should have a system that is ready for certification,’ Agbana said. ‘Our partners in the field have collected about 800 samples now. We hope to test them with the schistoscope and compare the results with gold standard methods.’
The microscope costs around €280, but Agbana hopes to reduce that to €80.
One-stop shop
While the products for miniature microscopy that Jenoptik offers are more expensive and sophisticated, they follow remarkably similar principles.
Ute Hofmann, their product manager for biophotonics, said: ‘When we consider how we bring microscopy closer to the patient, I always talk about three key words: miniaturisation, digitalisation and automation.
‘Microscopy evolution over the past two decades has transformed the whole technology. We had those big compound microscopes, made to try things out. You can use different standard objectives.
‘They are big. They are heavy. With clever optical design, you can miniaturise them, make them lighter. You can squeeze down the size and you don’t need eye pieces anymore. You don’t need the camera itself. You just use the sensor. All these steps reduce the complex microscope to a small one.’
Jenoptik’s core business is putting different hardware and software components together to build small digital imaging solutions, which is the exact combination needed for miniature microscopes. ‘Usually, life science instrument companies start to put together different optical components that were developed independently,’ Hofmann observed. ‘They might have the wrong camera. The sensor doesn’t fit with the illumination or with the objective lens performance. Our offer: we align all these different optical components together so that every photon that is necessary to count is delivered from the sample to sensor. We develop the optical subsystems that are necessary to enable the customers’ instruments.’
This philosophy is embodied in the Jenoptik Syions platform. ‘Jenoptik Syions has core technology with a PCB assembly electronic evaluation board and software subsystem,’ says Hofmann. ‘This is essential for all peripherals to join together. It is an evaluation kit with different illumination and detection modules.’
Jenoptik Syions offers companies developing new products a starting point that is ready to use, ‘but we always modify towards the application and the request from the customer’, Hofmann explained.

A four-channel fluorescence microscope including brightfield based on the Jenoptik Syions platform
Jenoptik Syions helps Jenoptik’s customers with lab prototypes that are too large and not cost effective. ‘With Jenoptik Syions we demonstrate that there is a much smaller solution than what they have at the moment, because we reduce complexity,’ Hofmann said.
‘With our configurable modules for light sources, optics, electronics, sensors and software that perfectly match, we create cutting-edge solutions for customers ready for flexible and seamless integration into their instruments. That means we do not start from scratch. As a one-stop-shop customers get everything from one single source with us.’
In haematology, Jenoptik and its partners are exploiting this concept several steps after a first stage of counting numbers of blood cells using flow cytometers. ‘At the very end of the line in haematology is microscopic observation,’ said Hofmann. ‘Usually this is still done with the standard microscope and not automated. We’re working on a project where we try to automate this image processing as well, for detection of circulating tumour cells.’
Recreating and then recognising images
While such projects focus on making best use of existing technology, researchers at CEA Leti in Grenoble, France, have developed an innovative approach to miniaturise microscopy further, with Lensfree.
Sophie Morales, their head of imaging systems for biology and health, said: ‘The system operates with a near-infrared light emitted by an LED that is diffracted by the biological sample being analysed to generate a holographic pattern captured by a CMOS image sensor,’ she said. ‘Holographic reconstruction algorithms digitally recreate the image of the object on a display. AI software then detects, analyses, and even classifies biological objects by tracking metrics of interest. All of these steps are automated.’
Morales defines the approach as computational microscopy, because an algorithm replaces the lenses used in traditional microscopes. ‘The hardware is generic for all the applications and we have developed a variety of dedicated algorithms to address a very large amount of different applications. It is really powerful and simple to use,’ she said.
‘You just have to put your sample on top of the CMOS sensor and everything can be automatic. The very simple hardware architecture of Lensfree allows the miniaturisation of equipment dedicated to in-vitro diagnostics, miniaturisation that opens the doors to patient diagnosis outside the lab. The set-up is very small, since all the lenses have been removed and several point-of-care systems at the bedside of the patient can now be imagined.’

CEA Leti’s Lensfree technology enables microscopes that use holographic reconstruction algorithms to digitally recreate the image of an object on a display from diffracted raw data, before AI software detects, analyses and classifies biological objects
CEA-Leti has partnered with several organisations to bring the technology into practical use. Some of them just sell Lensfree instruments, while for others the business model is to sell the instrument cheaply, and generate revenue from related consumable items. For example, Leti has applied the technology to complete blood count processes in partnership with Horiba Medical, in Montpellier, France, over a six-year period. ‘We are at the end of the process of research and development,’ said Morales. ‘We hope that a product will soon be commercialised by Horiba.’
CEA-Leti has similarly worked with Avalun, a startup in Grenoble, France, to produce a digital blood coagulation test. With Marseille-Méditerranée University Medical Centre, CEA-Leti has also developed a point-of-care system for meningitis detection using spinal fluid.
Agbana stressed the importance that specialised miniature microscopes can have. ‘Go to the villages,’ he said, hoping to reach partners who might also like to work on the schistoscope project. ‘Look at the real conditions. These are neglected tropical diseases that have a long-term impact, particularly on children. We can touch more lives together, we can impact more people. And we can solve a problem that can redefine the destiny of a child.’
Functional genomics screening with laser-based light engines
By supplying approximately one watt of optical power from each of its seven laser sources, the Celesta light engine is well-suited to high-content screening
High-content screening (HCS) combines automated fluorescence microscopy and quantitative data analysis in a high-throughput format suitable for large-scale applications such as drug discovery and systems biology. The usefulness of HCS derives from the information it provides on cellular phenotypes and how they respond to pharmacological or genomic manipulations. The range of detectable responses can be much broader than accessed by simple biochemical assays, hence the appellation ‘high-content’.
The recent publication by Wheeler and co-workers1 that forms the basis of this case study illustrates two noteworthy advances in HCS implementation:
- Use of three-dimensional (rather than two-dimensional) fluorescence microscopy
- Use of cellular arrays in which groups of cells can be cultured separately, and from which cells presenting novel or interesting phenotypes can be selectively removed for downstream genomic or proteomic analysis
The motivation for the use of three-dimensional microscopy is increased spatial resolution. In HCS, the objective of the analysis is often to determine whether two components of a cell are adjacent to each other, in which case they can interact and produce a response, or not.
In the simplistic depiction shown in figure 1, the red and blue disks, representing components of a spherical cell, appear to be coincident in the two-dimensional view, but are shown to be separated when viewed in three dimensions. In the vernacular of HCS, the two-dimensional view is a false positive result. The motivation for post-hoc genomic or proteomic analysis is to expand on the phenotypic information provided by fluorescence microscopy. In turn, this expansion of scope has been driven by the development of genomic manipulation tools such as Crispr editing. Crispr is a family of genome sequences.
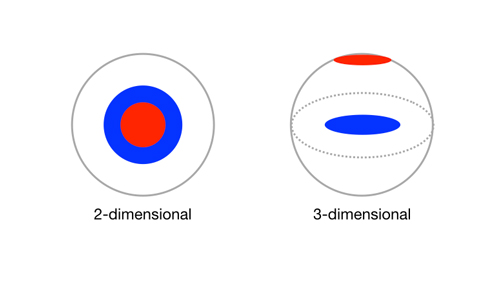
Figure 1: Schematic representations of two non-adjacent cellular components (red and blue disks) viewed in two and three dimensions
Wheeler and co-workers1 note that integration of pooled Crispr genetic screening with cellular and subcellular imaging readouts is critical to improving phenotypic definition in image-based genetic knockout studies. They describe screening Crispr-infected HEK293T cells on microraft arrays, followed by automated high-resolution confocal imaging to identify regulators of stress granules, which are punctate protein–RNA assemblies that form during cellular stress. The screen identified and validated six previously known stress granule modulators, along with 17 RNA-binding proteins (RBPs) that, when depleted, reduce sodium arsenite-induced stress granules in human cells.
Microraft arrays (commercially available from Cell Microsystems; Durham, NC; figure 2) are an attractive platform for screening bulk-infected cells because thousands of clonal cell colonies (~5 to 20 cells per colony) can be cultured in isolation from one another. Three-dimensional imaging of microraft arrays was accomplished using a Celesta light engine coupled to a CrestOptics X-Light V2 L-FOV spinning disk confocal scanner (figure 3). The 405, 520, 546 and 638nm output lines of the Lumencor Celesta light engine were used to excite fluorescence of DAPI, mCitrine, mCherry and Alexa Fluor 633 respectively.

Figure 2: Cellular phenotypes displayed on a 6 x 5 microraft array. Each 100μm square raft consists of magnetic polystyrene embedded in a polydimethylsiloxane (PDMS) substrate
Several attributes of the laser-based Celesta light engine are central to its use in this application. Firstly, it delivers the spectral content required for independently controlled excitation of the four fluorescent markers from which the image data is generated. Secondly, the increased spatial resolution of three-dimensional imaging has a cost in terms of diminished light throughput. The Celesta light engine compensates for this diminished throughput by supplying approximately one watt of optical power from each of its seven laser sources.
Finally, although the imaging scale is microscopic, the instrumentation required to execute automated three-dimensional imaging of living cells is decidedly macroscopic, occupying a considerable amount of laboratory space. The compact footprint and capacity for remote control of the Celesta light engine allows it to be installed unobtrusively, up to six feet away from the microscope body (figure 3); a considerably more stable and robust illumination platform than a laser table outfitted with all the sources and optics required for such multi-parameter analyses.

Figure 3: Instrumentation for automated three-dimensional imaging of living cells. The system is a Nikon Ti2 inverted microscope equipped with an environmental enclosure for maintenance of cellular homeostasis. On the right is an incubator for storage of live cell arrays, before and after analysis. To the rear, left of the microscope is the Lumencor Celesta light engine
Reference
[1] EC Wheeler, AQ Vu, GW Yeo et al. Nature Methods (2020) 17: 636–642


