Microendoscopy, or ‘optical biopsy’, combines the use of optical fibres with microscopy techniques such as optical coherence tomography (OCT) to image tissues inside the human body, in vivo. Clinicians hope to use this to diagnose diseased tissues without the need for invasive biopsies.
Microendoscope technology is relatively new, with many advances having been made only in the past decade. A handful of companies currently offer clinically approved microendoscopy solutions: the FIVE2 from OptiScan Imaging in Australia; Convivo, the neurosurgical counterpart from Carl Zeiss Meditech in Germany; and the Cellvizio from Mauna Kea Technologies in France.
The low cut-through of fibre-based microendoscopy despite increasing clinical need is in part due to steep physical limitations. These limitations arise from the need for a long working distance (or focal length), as well as for high optical resolutions and increasingly small probe diameters.
These shortcomings result in the technology struggling to compete with bulk microscopes. Confocal endomicroscopy, for example, can provide the necessary resolution to visualise tissues and provide simple diagnosis at the cellular level, but cannot achieve the penetration depth and field of view for impact in clinical settings. Other competing technologies, such as conventional optical coherent tomography (OCT), provide better imaging depth and larger fields of view, but cannot achieve high resolutions or require bulky probes. These trade-offs have so far prevented microendoscopes from revolutionising clinical diagnostics, but researchers are paving the way to better techniques.
Digging deeper: two photons or three?
To target higher resolutions with better penetration, researchers have in recent years moved away from standard confocal microscopy techniques, instead looking at two-photon and three-photon imaging methods. In these techniques, an ultrafast pump laser is used to generate two or three photon excitations in a biological sample, with the resulting fluorescence imaged through an endoscopic probe.
“You have natural confocality [with two-photon microscopy] because you have two-photon emission only at the focal spot. Thanks to that, you can go deeper into [say] the brain, as far as a few hundred microns,” explains Alexandre Kudlinski, a fibre-optics researcher based at the University of Lille and a founder of LightCore Technologies, which markets the BondXplorer Microscope, a multimodal endoscope based on two-photon microscopy.
Scientists such as Kudlinski are hoping to open a new sphere of applications in microendoscopy: real-time in vivo imaging. They are specifically targeting the imaging of neural networks in mouse brains. Currently, mouse brains are examined under bulk microscopes where the mice must be kept still to remain under the microscope objective. Kudlinski and his collaborators at both Lille and Lightcore are working on a plug-and-play endoscope that can be inserted into a helmet worn by a mouse.
“Because the endoscope is flexible, the mouse can move. It can eat, it can drink, it can play, it can fight with its sister,” says Kudlinski. “The neuroscientists can study neuronal activity while the mouse is freely moving and doing all these activities. If you have transgenic mice then you can monitor brain activity in real time. With our endoscope, this is possible.” Kudlinski has collaborated on developing microendoscope probes with specialists in bio-imaging from the Universities of Limoges and Marseille. He and his collaborators spun out Lightcore Technology in 2019, where they have moved on to developing microendoscopic systems. Lightcore sells two-photon and threephoton microendoscope systems based on commercially available laser systems. For two-photon imaging, it employs standard titanium sapphire systems operating at 80MHz with about 150 femtosecond (fs) pulse durations, either at 800nm or 920nm to suit the green fluorescent protein used as a biological marker in neuroscience.
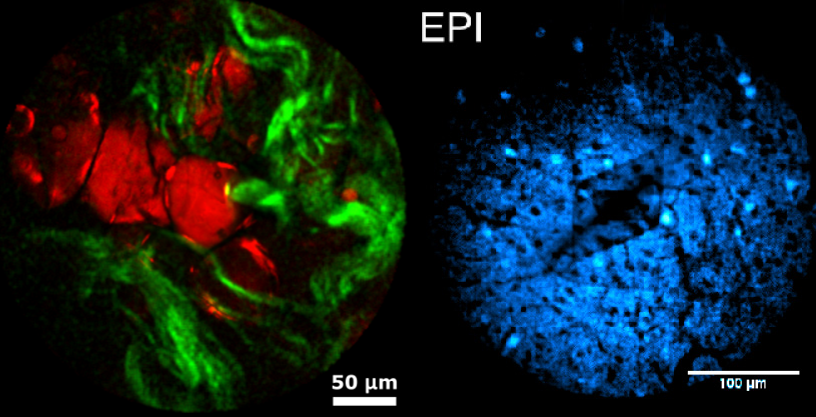
Left: InSplorer shots of human colon cells, with lipids (red) and collagen (green). Right: Two-photon image of a mouse brain labelled with GCaMP indicator (Image: LightCore Technologies)
For three-photon processes, shorter pulses (less than 50fs) are required to generate the relevant peak powers. To do this, they must go to longer wavelengths, such as 1,300nm or 1,700nm, to suit the biological markers. This is typically done with ytterbium-doped fibre oscillator systems pumping large bulk optical parametric amplifiers (OPAs) that can be tuned, for example, around 1,300nm. Lightcore Technologies, in its InSplorer Endoscope module, uses a combination of Coherent’s Monaco as the ytterbium-based pump laser, with an Opera OPA. The use of three-photon microscopy enables higher penetration depths than possible with two-photon techniques, as well as increased resolution.
“If you want to target the same [biological] marker, you use a higher wavelength such as 1,300nm or 1,700nm. At these wavelengths, water absorption is reduced in the tissues, so you can penetrate deeper,” says Kudlinski. “The advantage of three-photon is that you have a better actual resolution. The transverse resolution is the same, but the longitudinal sectioning is improved with three-photon imaging. So, for a 3D image you get a better resolution in terms of depth.”
Hollowcore fibre is also a key piece of the puzzle for two- and three-photon imaging, because of the ultrashort pulse durations required.
“If you use solid core fibre, then you need to pre-compensate for dispersion and nonlinearity,” says Kudlinski. “In hollowcore fibre, intense pulses are propagating in air, so the nonlinearity is not such a problem. The group velocity is also very, very small, so it doesn’t need to be pre-compensated. For a 40fs pulse at 800nm, it can be propagated in hollowcore fibre for two or three metres without temporal broadening.”
Two-photon and multiphoton endoscopes are not currently available to clinicians, but the technology is steadily developing. Multiphoton microendoscopes are “not far” from clinical readiness, says Kudlinski. “It’s mainly linked to the security around the endoscope. Everything needs to be bio-compatible and right now we have a laboratory prototype. But from a technological point of view, we are very close.”
Kudlinski is cautious to note, however, that multiphoton microendoscopes are not a hero technology. Bulk microscopes still offer better performances in terms of field of view, offering several millimetres where fibre-based probes can only offer a few hundred microns. There are also limitations in nonlinear microendoscopy due to available penetration depth, because of miniaturisation of the device. The working distance is much smaller than in bulk microscopes because of the focusing optics used. Reaching the performance of a bulk microscope in terms of field of view would be “quite tricky” with current focusing technologies.
“You have to find a compromise between the size of the device – of the endoscopic head – and the optical performances. There is no trick for that,” says Kudlinski.
Moving towards the miniature
As Kudlinski notes, trade-offs are usually made in microendoscope systems between imaging resolution and device size based on the focusing elements used to collect light in the endoscope probe. Typical designs are based on one of two types of focusing elements: graded-index (GRIN) fibre probes (GFPs), or spherical ball lens probes (BLPs).
Both GFPs and BLPs have their advantages and disadvantages. GFPs are easy to fabricate and are robust to the refractive indices of the imaging medium, but high resolution is hard to achieve with commercially available GRIN fibres with small core diameters, and these probes suffer from severe astigmatism in sideviewing configurations. Spherical BLPs, on the other hand, do not suffer from astigmatism, but have a heavy dependence on the refractive index of the imaging media, making them difficult to use in biological materials. They are also bulky, which complicates miniaturisation efforts. Multi-element systems combining multiple lenses could provide a limited solution, but are relatively difficult to manufacture.
However, innovations have been made in recent years to overcome the trade-offs faced by microendoscope systems. A 2022 study in IEEE Photonics Journal, led by Karol Karnowski of Poland’s International Centre for Translational Eye Research, has developed a new type of microendoscopic probe that “pave[s] the way for a broader range of imaging applications” by offering enhanced optical performance at different working distances.
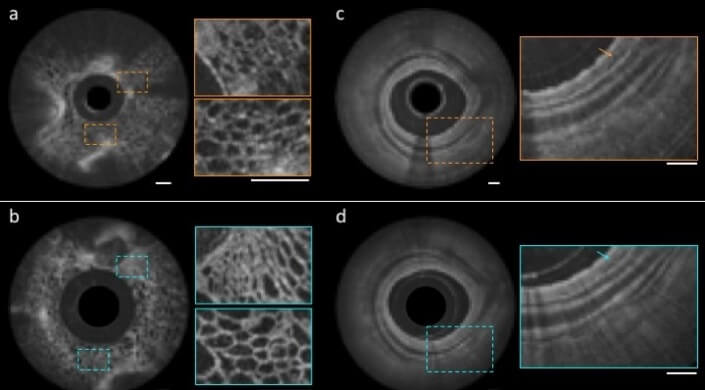
An image of a grape taken using a conventional OCT catheter (a) and with a nano-optic, metalens-equipped endoscope (b); and of a swine airway using a ball lens catheter (c) and nano-optic endoscope (d). The scale bars shown are 500µm
Karnowski and co-authors have developed a new focusing method in their endoscopic probes. By combining both GRIN fibres and SBLs, they show “superior performance over a range of numerical apertures”. Their results have been particularly useful in the miniaturisation of microendoscopic probes, showing that their probes can achieve the same or better imaging performance as single-element probes twice their diameter.
“Previously, we’ve used probes made from GRIN lenses of 1mm diameters, with a focal length of 5mm or 6mm. This gives a tubing of 3mm outer diameter,” says Karnowski. “With the GRIN fibre and ball lens configuration, you can half this. If you think of inserting a 3mm tube into your airway, compared with a probe half this size, then the advantage is obvious.
Karnowski adds that the combination also offers both better resolution “and, if you manage to make the ball lens from a higher refractive index material, there is a possibility of higher focusing power, meaning a short working distance. In combination with GRIN fibre, you could possibly push the working distance even further.”
In the combined GRIN fibre and ball lens configuration, the issues of astigmatism faced using purely GRIN fibre are solved. The technology is also robust in varying imaging media, meaning that new applications could open up where index matching is not required between probe and sample.
Wide-scale application and low cost are important to Karnowski: “I have a drive around the simplification of things, to make things available for many people. One parameter is price. My dream would be to propose a way to fabricate probes that would be cheap. It all starts with research.” The team says they were driven by a desire for simplicity, low cost, and repeatability and, as a consequence, the combined probes are easy to manufacture. They use a combination of single-mode fibre, GRIN fibre and coreless spacers, applying heat to create a ball lens. “In the time you take to make one probe from GRIN lenses, you can make a few or even tens of probes from GRIN fibre optics, and with modern splicers, you have a great deal of control,” says Karnowski. “For GRIN lens [probes], you have to glue fibres to a ferrule, wait for curing and polishing, glue and then polish the lens to the correct focal length. If you fail on the last process, you’re done.”
A new scope: the future of microendoscopes
Karnowski’s group is not the only one pushing the boundaries, and other technologies are finding routes to circumvent the issues of miniaturisation, working distance and resolution. In 2018, a research group at Harvard John A. Paulson School of Engineering and Applied Sciences applied metalenses to develop a new type of nano-endoscope, with the potential to overcome imaging limitations.
Described as “game-changing” by Federico Capasso, one of the lead principal investigators on the study, the metalens probe is based on endoscopic OCT. The group used metasurfaces, two-dimensional metamaterials that can flexibly control the properties of light, to overcome barriers to excellent focusing. This could potentially enable high-quality imaging in complex media with arbitrary refractive indices, such as the human body.
More recently, researchers at the Universities of Stuttgart and Adelaide used 3D printing to produce the world’s smallest endoscope, with a width of just 125 microns. The work was presented in Light: Science and Applications in a study led by Jiawen Li. 3D-printed elements have the potential to push the miniaturisation of endoscopes far past current limits, but the technology remains in its infancy.
Dr Anita Chandran is a writer based in Hamburg, Germany. She has a PhD in ultrafast fibre lasers and has worked in historical fiction, science pedagogy and AI ethics.
--
Sponsored Case Study: Functional genomics screening with laser-based light engines
By supplying approximately one watt of optical power from each of its seven laser sources, the CELESTA light engine is well-suited to high-content screening.
High-content screening (HCS) combines automated fluorescence microscopy and quantitative data analysis in a high-throughput format suitable for large-scale applications such as drug discovery and systems biology. The usefulness of HCS derives from the information it provides on cellular phenotypes and how they respond to pharmacological or genomic manipulations. The range of detectable responses can be much broader than accessed by simple biochemical assays, hence the appellation ‘high-content’.
The recent publication by Wheeler and co-workers1 that forms the basis of this case study illustrates two noteworthy advances in HCS implementation:
-
Use of three-dimensional (rather than two-dimensional) fluorescence microscopy
-
Use of cellular arrays in which groups of cells can be cultured separately, and from which cells presenting novel or interesting phenotypes can be selectively removed for downstream genomic or proteomic analysis.
The motivation for the use of three-dimensional microscopy is increased spatial resolution. In HCS, the objective of the analysis is often to determine whether two components of a cell are adjacent to each other, in which case they can interact and produce a response, or not.
In the simplistic depiction shown in Figure 1, the red and blue disks, representing components of a spherical cell, appear to be coincident in the two-dimensional view, but are shown to be separated when viewed in three dimensions. In the vernacular of HCS, the two-dimensional view is a false positive result. The motivation for post-hoc genomic or proteomic analysis is to expand on the phenotypic information provided by fluorescence microscopy. In turn, this expansion of scope has been driven by the development of genomic manipulation tools such as CRISPR editing. CRISPR is a family of genome sequences.
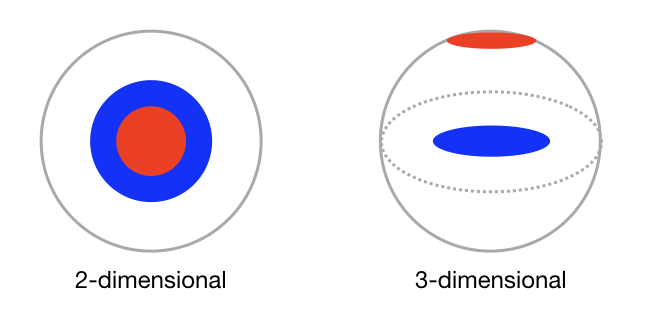
Figure 1: Schematic representations of two non-adjacent cellular components (red and blue disks) viewed in two and three dimensions
Wheeler and co-workers1 note that integration of pooled CRISPR genetic screening with cellular and subcellular imaging readouts is critical to improving phenotypic definition in image-based genetic knockout studies. They describe screening CRISPR-infected HEK293T cells on microraft arrays, followed by automated high-resolution confocal imaging to identify regulators of stress granules, which are punctate protein-RNA assemblies that form during cellular stress. The screen identified and validated six previously known stress granule modulators, along with 17 RNAbinding proteins (RBPs) that, when depleted, reduce sodium arsenite-induced stress granules in human cells.
Microraft arrays (commercially available from Cell Microsystems; Durham, NC; Figure 2) are an attractive platform for screening bulk-infected cells because thousands of clonal cell colonies (~5 to 20 cells per colony) can be cultured in isolation from one another. Three-dimensional imaging of microraft arrays was accomplished using a CELESTA light engine coupled to a CrestOptics X-Light V2 L-FOV spinning disk confocal scanner (Figure 3). The 405, 520, 546 and 638nm output lines of the Lumencor CELESTA light engine were used to excite fluorescence of DAPI, mCitrine, mCherry and Alexa Fluor 633 respectively.

Figure 2: Cellular phenotypes displayed on a 6x5 microraft array. Each 100μm square raft consists of magnetic polystyrene embedded in a polydimethylsiloxane (PDMS) substrate
Several attributes of the laser-based CELESTA light engine are central to its use in this application. Firstly, it delivers the spectral content required for independently controlled excitation of the four fluorescent markers from which the image data is generated. Secondly, the increased spatial resolution of three-dimensional imaging has a cost in terms of diminished light throughput. The CELESTA light engine compensates for this diminished throughput by supplying approximately one watt of optical power from each of its seven laser sources.
Finally, although the imaging scale is microscopic, the instrumentation required to execute automated three-dimensional imaging of living cells is decidedly macroscopic, occupying a considerable amount of laboratory space. The compact footprint and capacity for remote control of the CELESTA light engine allows it to be installed unobtrusively, up to six feet away from the microscope body (Figure 3); a considerably more stable and robust illumination platform than a laser table outfitted with all the sources and optics required for such multiparameter analyses.

Figure 3: Instrumentation for automated three-dimensional imaging of living cells. The system is a Nikon Ti2 inverted microscope equipped with an environmental enclosure for maintenance of cellular homeostasis. On the right is an incubator for storage of live cell arrays, before and after analysis. To the rear, left of the microscope is the Lumencor CELESTA light engine
For more information visit: https://lumencor.com
Reference
[1] EC Wheeler, AQ Vu, GW Yeo et al. Nature Methods (2020) 17: 636–642


